Garbage In, Garbage Out: Data Quality is Essential for RNA Biomarker Discovery
At Decode Health, we empower the healthcare industry to implement and leverage artificial intelligence (AI) approaches, including machine learning, across a wide range of clinical applications. Data quality is paramount, especially when utilizing large, disparate datasets to address complex diseases. Decode has developed a platform that ensures the precision and reproducibility of insights and results from diverse data sources. The platform features a robust quality control (QC) framework that optimizes the efficiency and accuracy of biomarker discovery. Check out the article below to learn how our technology accelerates the biomarker discovery R&D process, widens the impact of personalized medicine, and unlocks new insights to fuel better ways to diagnose and treat patients.
In the evolving field of precision medicine, Decode Health has developed an advanced analytics platform leveraging artificial intelligence (AI) methods, including machine learning (ML), to efficiently identify candidate biomarkers for acute and chronic conditions. We have integrated a robust quality control (QC) system within this platform to facilitate fast, precise, and reproducible outcomes during the biomarker discovery process. The QC framework includes thorough quality checks to ensure a high level of result integrity and reliability. Attention to specific quality measurements at each stage of the process permits the identification of reliable biomarkers across a wide range of complex diseases, including autoimmune disorders, inflammatory and non-inflammatory neurologic conditions, and metabolic disorders.
Decode has developed a unique quality control framework that ensures quality throughout the entire end-to-end process, from the pre-analytical to the analytical stages. This framework is designed to identify and address poor-quality inputs as early as sample collection. Variations in sample handling can negatively impact the quality of inputs, which can lead to inaccurate results and reduce the reproducibility of biomarker discovery programs. This is especially important for programs that use RNA sequencing, where adherence to uniform protocols is essential to ensure sample stability and isolation of high-quality nucleic acids. The Decode QC framework has been developed from internal and external best practices and applied to hundreds of clinical specimens covering various clinical conditions such as multiple sclerosis (MS), neuromyelitis optica (NMO), insulin sensitive, insulin resistant, and healthy individuals. Decode has optimized laboratory and analytical processes to improve the clarity of results, emphasizing the need for quality inputs to ensure quality outputs and adapting the QC framework accordingly. Decode’s platform delivers candidate biomarkers faster than traditional discovery approaches, and the embedded QC framework provides increased confidence in the identified candidate biomarkers. This balance is crucial in the fast-paced world of medical research, where the speed of discovery is often as important as the accuracy of the findings. Decode’s approach significantly accelerates and improves the successful transition from research to translational, real-world applications.
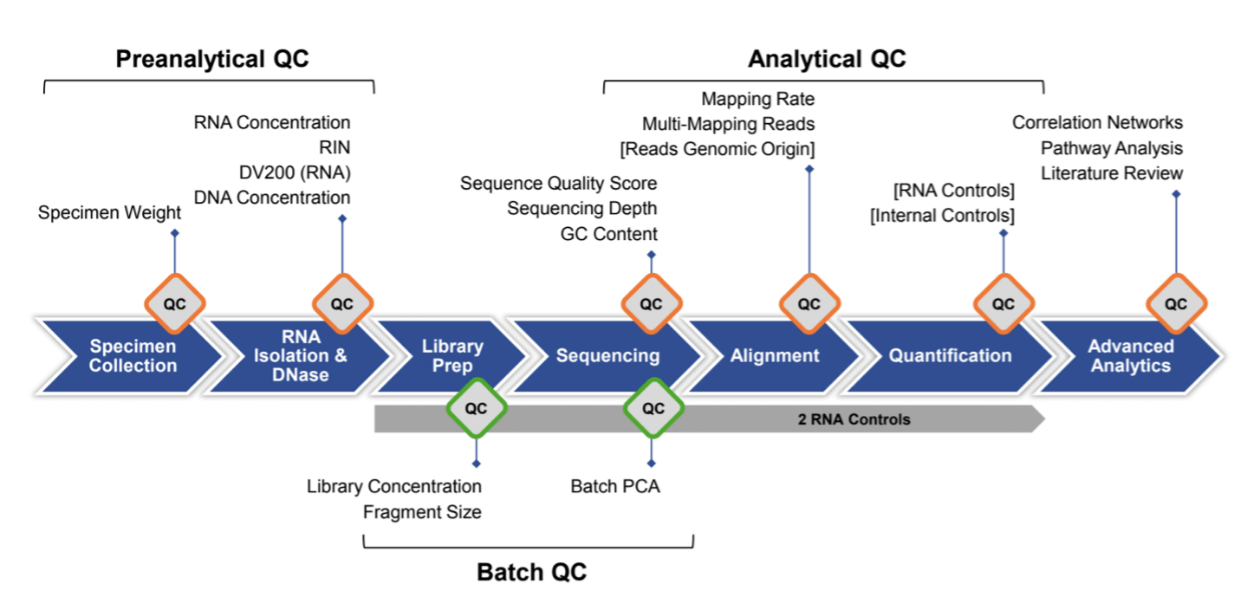
Preanalytical and Analytical Quality Control Diagram Demonstrating the End-to-End Framework
Example workflow diagram of quality control (QC) checkpoints and metrics evaluated for whole blood specimens. RNA controls are integrated before library preparation. Orange QC checkpoints indicate metrics assessed on individual samples, while green QC checkpoints indicate metrics assessed on batches of samples. Brackets indicate parameters currently under development. RIN, RNA integrity number; DV200 (RNA), percentage of RNA fragments > 200 nucleotides; PCA, principal component analysis
Decode’s advanced analytics platform enables faster and more reliable biomarker discovery, leading to improved diagnostics and personalized treatment strategies. In the field of precision medicine, identifying candidate biomarkers can enhance the diagnosis or subtyping of complex diseases and uncover new therapeutic targets. At the 2023 Association for Molecular Pathology annual meeting, Decode presented work highlighting the QC framework and applications of our AI-enabled advanced analytics platform using RNA sequencing in metabolic and neurodegenerative conditions. We demonstrated that:
- Focusing on quality in an end-to-end pipeline for RNA biomarker discovery begins at specimen collection. With many years of experience in RNA diagnostics, Decode collaborated with Quest Diagnostics to establish a nationwide biospecimen collection initiative to minimize RNA quality issues during specimen collection.
- Sample and data quality issues can be identified and addressed early in the biomarker discovery pipeline.
- The QC framework is adaptable to multiple specimen types, including whole blood and plasma. However, pre-analytical assessments for RNA integrity and concentration must be carefully considered before applying advanced analytics and introducing unintended biases.
- The platform can be applied to gain insights and identify prospective biomarkers in smaller, early-stage proof-of-concept studies and larger, validation studies to power novel, biologically-relevant biomarker findings.
- a proof-of-concept metabolic study, prospective RNA biomarkers for insulin resistance could be identified in whole blood and plasma samples. These RNA biomarkers were associated with critical and metabolically relevant biological processes, including N-glycan synthesis, cellular senescence, and coagulation factor activity.
- In our analysis of neuroinflammatory disease, triangulation of multiple methods identified a robust set of candidate RNA biomarkers distinguishing MS from NMO and healthy controls. These biomarkers are associated with biological pathways, including bacterial responses, membrane pore formation, and regulation of hemoglobin synthesis. Analysis of previously published scientific literature reinforced the association of these pathways with inflammatory neurologic conditions. Through this process, we identified candidate biomarkers for disease diagnostics and potential therapeutic targets.
It is well-known and documented that poor-quality data inputs lead to poor reproducibility in biomarker identification studies. Decode’s data platform with an embedded, comprehensive QC framework includes safeguards that mitigate common pitfalls in the biomarker discovery process, particularly for RNA sequencing applications. Decode presents these concepts at national and international conferences to promote greater collaboration and standardization of biomarker discovery practices and ensure future data sharing and larger meta-analyses are not subject to bias or technical issues. Our recent presentations show how our technology and technical approach can be harnessed to overcome common research barriers that delay or prevent the discovery of personalized healthcare solutions.
To learn more, email Decode Health at info@decodehealth.ai